In the Lewis Group
Enzymes are remarkable catalysts that have the potential to greatly improve the efficiency of chemical synthesis. They exhibit extremely high catalytic proficiency and selectivity toward their native substrates, and they typically operate at ambient temperature and pressure within narrow pH and redox potential ranges in aqueous solution. New enzymes are constantly being identified from studies on natural product biosynthesis, genome mining, de novo design, and other approaches. Unfortunately, however, the enzymes obtained from these efforts rarely possess useful levels of activity or selectivity on substrates of interest, and their activity is often compromised under preparative reaction conditions (high concentration, organic co-solvents, etc.). Central to any effort to employ enzymes for chemical synthesis is a robust method to improve their catalytic properties. In the Lewis group, we use directed evolution for this purpose:
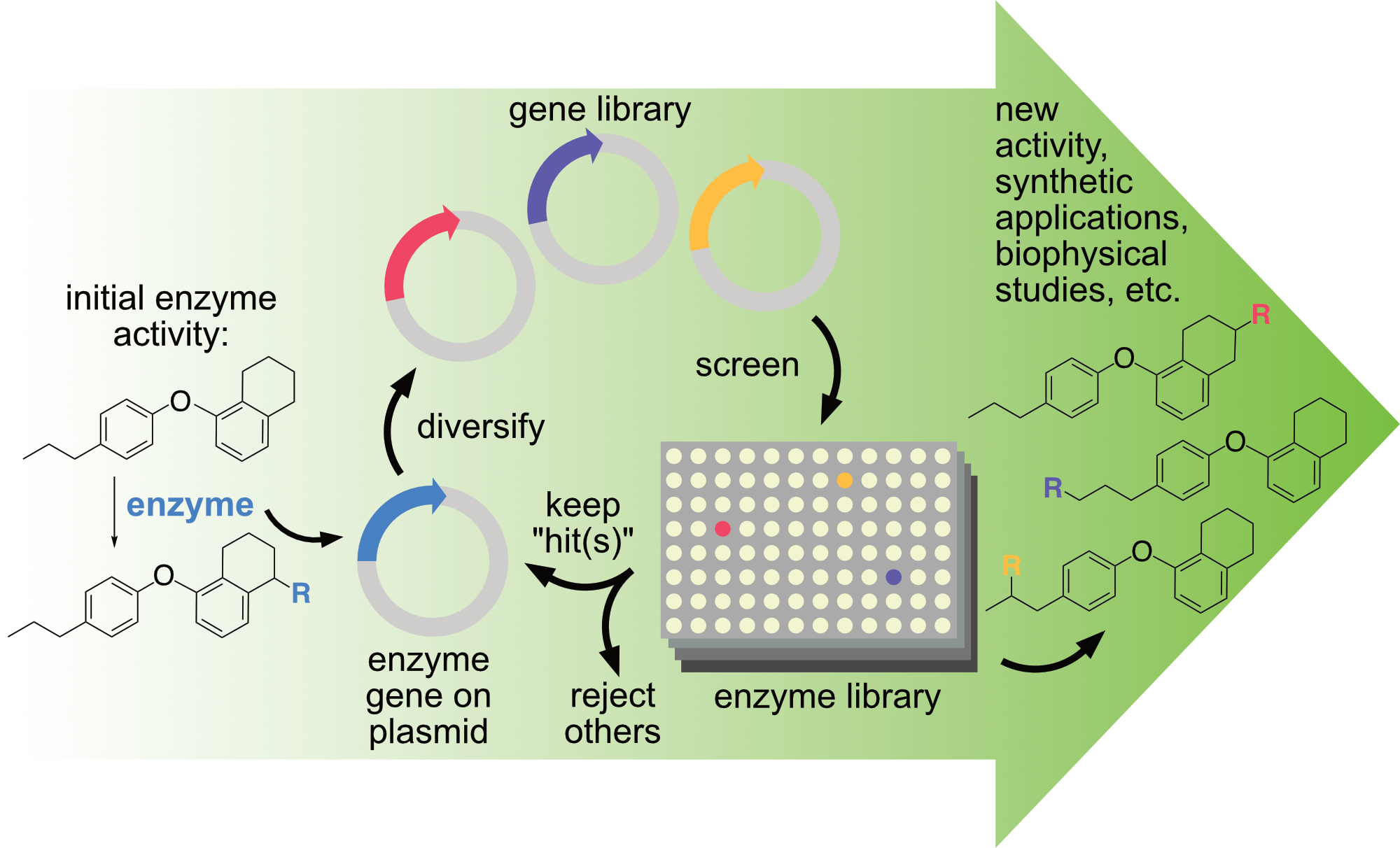
At its core, directed evolution involves diversifying a gene encoding an enzyme of interest, screening the activity of the enzyme variants, and repeating the process on the gene(s) encoding improved variants while rejecting others. In this way, the native activity and selectivity of an enzyme of interest can be rapidly altered or improved to enable new reactions under conditions suitable for preparative reactions. This approach lies at the heart of nearly all projects in the Lewis group. Whether students are working with natural enzymes or artificial metalloenzymes, they have the opportunity to learn state-of-the-art techniques in directed enzyme evolution. Most projects also leverage an integrated robotic platform to automate nearly every step of directed evolution. This system was constructed using funds from a Defense University Research Instrumentation Program (DURIP) award from the U.S. Army Research Office (66796-LS-RIP). It is compatible with enzymes that catalyze a wide range of reactions and provides facile access to library sizes of consisting of thousands variants. See the system in action here:
We are also developing new approaches to access much larger library sizes using techniques such as continuous evolution and fluorescence activated cell sorting. While these approaches are more limited in the range of compounds and reactions to which they can be applied, they have the potential to dramatically accelerate the evolution process and thus allow us to probe different questions regarding enzyme evolution and catalysis.
Background
As noted above, directed evolution of proteins involves iterative mutation of a protein and functional screening/selection of the resulting variants to create protein variants with improved properties. Prior to the development of directed evolution, protein engineering, such as it was, largely involved targeted point mutation of proteins and thus only modest improvements in function. Directed evolution revolutionized this process and has led to stunning improvements in protein function for fine/commodity chemical synthesis, selective catalysis, pharmaceuticals development, materials science, chemical biology, and a wide range of other fields. It is safe to say that any application that uses a protein could benefit from some form of directed evolution at some point in its development. Moreover, directed evolution has influenced the means by which protein function and natural evolution is understood and studied. Two seminal and complementary methods, one by Prof. Frances Arnold published in 1993 and one by Dr. Willemm (Pim) Stemmer (deceased) published in 1994, mark the advent of directed evolution, and the pair received the 2011 Draper Prize for their discoveries. One half of the 2018 Nobel Prize in Chemistry was awarded to Prof. Arnold for her ground-breaking work on “the directed evolution of enzymes”. Prof. Lewis was a postdoctoral fellow with Prof. Arnold at Caltech from 2008-2010.
